Pasados 3 meses,los anticuerpos parecen desaparecer no así la inmunidad
Observación longitudinal y disminución de las respuestas de los anticuerpos neutralizantes en los tres meses siguientes a la infección por SARS-CoV-2 en humanos
Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans
Nature Microbiology (2020)
Abstract
Antibody responses to SARS-CoV-2 can be detected in most infected individuals 10–15 d after the onset of COVID-19 symptoms. However, due to the recent emergence of SARS-CoV-2 in the human population, it is not known how long antibody responses will be maintained or whether they will provide protection from reinfection. Using sequential serum samples collected up to 94 d post onset of symptoms (POS) from 65 individuals with real-time quantitative PCR-confirmed SARS-CoV-2 infection, we show seroconversion (immunoglobulin (Ig)M, IgA, IgG) in >95% of cases and neutralizing antibody responses when sampled beyond 8 d POS. We show that the kinetics of the neutralizing antibody response is typical of an acute viral infection, with declining neutralizing antibody titres observed after an initial peak, and that the magnitude of this peak is dependent on disease severity. Although some individuals with high peak infective dose (ID50 > 10,000) maintained neutralizing antibody titres >1,000 at >60 d POS, some with lower peak ID50 had neutralizing antibody titres approaching baseline within the follow-up period. A similar decline in neutralizing antibody titres was observed in a cohort of 31 seropositive healthcare workers. The present study has important implications when considering widespread serological testing and antibody protection against reinfection with SARS-CoV-2, and may suggest that vaccine boosters are required to provide long-lasting protection.
Las respuestas de los anticuerpos al SARS-CoV-2 pueden detectarse en la mayoría de los individuos infectados entre 10 y 15 d después de la aparición de los síntomas del COVID-19. Sin embargo, debido a la reciente aparición del SARS-CoV-2 en la población humana, no se sabe por cuánto tiempo se mantendrán las respuestas de los anticuerpos o si proporcionarán protección contra la reinfección. Utilizando muestras de suero secuenciales recogidas hasta 94 d después del inicio de los síntomas (POS) de 65 individuos con infección de SARS-CoV-2 confirmada por PCR en tiempo real, mostramos la seroconversión (inmunoglobulina (Ig)M, IgA, IgG) en >95% de los casos y la neutralización de las respuestas de los anticuerpos cuando la muestra supera los 8 d POS. Mostramos que la cinética de la respuesta de los anticuerpos neutralizantes es típica de una infección viral aguda, con títulos decrecientes de anticuerpos neutralizantes observados después de un pico inicial, y que la magnitud de este pico depende de la gravedad de la enfermedad. Aunque algunos individuos con una dosis infecciosa de pico alto (ID50 > 10.000) mantuvieron títulos de anticuerpos neutralizantes >1.000 a un POS >60 d, algunos con un pico ID50 más bajo tuvieron títulos de anticuerpos neutralizantes que se acercaban a la línea de base dentro del período de seguimiento. Se observó un descenso similar en los títulos de anticuerpos neutralizantes en una cohorte de 31 trabajadores sanitarios seropositivos. El presente estudio tiene importantes repercusiones cuando se considera la posibilidad de generalizar las pruebas serológicas y la protección de los anticuerpos contra la reinfección con el SARS-CoV-2, y puede sugerir que se necesitan refuerzos de la vacuna para proporcionar una protección duradera.
Main
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a β-coronavirus responsible for coronavirus disease-19 (COVID-19). Spike (S) is the virally encoded surface glycoprotein facilitating angiotensin-converting enzyme-2 (ACE-2) receptor binding on target cells through its receptor-binding domain (RBD). In a rapidly evolving field, researchers have already shown that, in most cases, individuals with a confirmed PCR diagnosis of SARS-CoV-2 infection develop IgM, IgA and IgG against the virally encoded surface S glycoprotein and nucleoprotein (N) within 1–2 weeks post onset of symptoms (POS) and remain elevated after initial viral clearance1,2,3,4,5,6,7. S glycoprotein is the target for neutralizing antibodies, and a number of highly potent monoclonal antibodies have been isolated that predominantly target the RBD8,9,10,11. A wide range of SARS-CoV-2-neutralizing antibody titres has been reported after infection and these vary depending on the length of time from infection and the severity of disease3,4,5,12,13. Further knowledge on the magnitude, timing and longevity of neutralizing antibody responses after SARS-CoV-2 infection is vital for understanding the role neutralizing antibodies might play in disease clearance and protection from reinfection and/or disease. Furthermore, as huge emphasis has been placed on antibody reactivity assays to determine seroprevalence against SARS-CoV-2 in the community and estimating infection rates, it is important to understand immune responses after infection to define parameters in which antibody tests can provide meaningful data in the absence of PCR testing in population studies.
Antibody responses to other human coronaviruses have been reported to wane over time14,15,16,17. In particular, antibody responses targeting endemic human α- and β-coronaviruses can last for as little as 12 weeks18, whereas antibodies to SARS-CoV and MERS can be detected in some individuals 12–34 months after infection15,19. Cross-sectional studies in SARS-CoV-2-infected individuals have so far reported lower mean neutralizing antibody titres for serum samples collected at later timepoints POS (23–52 d)4,7,20. However, there is currently a paucity of information on the kinetics and longevity of the neutralizing antibody response using multiple sequential samples from individuals in the convalescent phase beyond 30–40 d POS3,5,21. The present study uses sequential samples from 65 individuals with PCR-confirmed SARS-CoV-2 infection and 31 seropositive healthcare workers (HCWs) up to 94 d POS to understand the kinetics of neutralizing antibody development, and the magnitude and durability of the neutralizing antibody response.
We demonstrate that the magnitude of the neutralizing antibody response is dependent on disease severity. In some individuals who develop modest neutralizing antibody titres after infection (ID50 (serum dilution that inhibits 50% infection) in the 100–300 range), titres become undetectable (ID50 < 50) or are approaching baseline after ~50 d, highlighting the transient nature of the neutralizing antibody response towards SARS-CoV-2 in some individuals. In contrast, individuals with high peak ID50 for neutralization maintain neutralizing antibody titres in the 1,000–3,500 range >60 d POS. The present study has important implications when considering protection against reinfection with SARS-CoV-2 and the durability of vaccine protection.
Results
Cohort description
The antibody response in 65 real-time quantitative (rtq)PCR-confirmed, SARS-CoV-2-infected individuals was studied over sequential timepoints. The cohort consisted of 59 individuals admitted to, and 6 HCWs at, Guy’s and St Thomas’ NHS Foundation Trust (GSTFT). The cohort were 78.5% male with an average age of 55.2 years (range 23–95 years) (Table 1). Information about ethnicity was not collected. A severity score was assigned to patients based on the maximal level of respiratory support required during their period of hospitalization. This cohort included the full breadth of COVID-19 severity, from asymptomatic infection to those requiring extracorporeal membrane oxygenation (ECMO) for severe respiratory failure. Comorbidities included diabetes mellitus, hypertension and obesity, with a full summary in Supplementary Table 1. Sequential samples were collected at timepoints between 1 and 94 d POS and were based on the availability of discarded serum taken as part of routine clinical care, or as part of a HCW study.
Antibody-binding responses to SARS-CoV-2
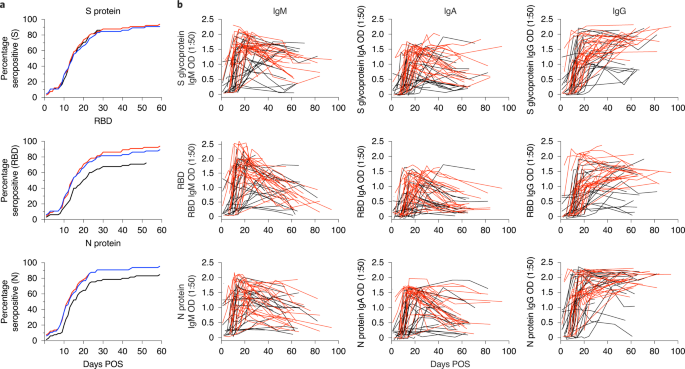
The IgG, IgM and IgA response against S glycoprotein, RBD and N protein were measured by enzyme-linked immunosorbent assay (ELISA) over multiple timepoints (Fig. 1 and Extended Data Fig. 1)6. Initially, the optical density (OD) at 1:50 serum dilution was measured for 302 samples from the 65 individuals (Fig. 1 and Extended Data Fig. 1). Only 2/65 individuals (3.1%) did not generate detectable antibody responses against these antigens in the follow-up period (Supplementary Table 2). However, sera were available only up until 2 and 8 d POS for these two individuals and, as the mean time to seroconversion against at least one antigen was 12.6 d POS, it is likely that these individuals may have seroconverted after they had been discharged from hospital. IgG responses against S, RBD and N antigens were observed in 92.3%, 89.2% and 93.8% of individuals, respectively (Supplementary Table 2). The frequency of individuals generating an IgM response was similar to that with IgG, with 92.3%, 92.3% and 95.4% seropositive against S glycoprotein, RBD and N protein, respectively. The frequency of individuals with an IgA response to RBD and N protein was lower, with only 72.3% and 84.6% seropositive, respectively (Supplementary Table 2), whereas the IgA to S glycoprotein frequency was similar to that of the IgM and IgG.
a, A cumulative frequency analysis describing the point of seroconversion for each individual in the cohort. Graph shows the percentage of individuals in the cohort who become IgM, IgA or IgG positive to S glycoprotein, RBD and N protein each day POS. A serum sample is considered positive when the OD is fourfold above background. b, OD values at 1:50 serum dilution for IgM, IgA and IgG against S glycoprotein, RBD and N protein over time. Each line represents one individual (n = 65). Severities 0–3 are shown in black and severities 4/5 in red. More than 300 pre-COVID-19 healthy control samples and >100 sera from PCR-confirmed SARS-CoV-2-infected individuals were previously used to develop and validate the ELISA setup6. The ELISAs were conducted once.
A cumulative frequency analysis of positive IgG, IgA and IgM responses against S glycoprotein, RBD and N protein across the cohort did not indicate a more rapid elicitation of IgM and IgA responses against a particular antigen (Fig. 1a and Extended Data Fig. 2a) and may reflect the sporadic nature in which serum samples were collected. Therefore, a subset of donors from whom sera were collected over sequential timepoints early in infection (<14 d POS) were analysed further and different patterns of seroconversion were observed (Extended Data Fig. 2b). Of these individuals, 51.6% (16/31) showed synchronous seroconversion to IgG, IgM and IgA, whereas some individuals showed singular seroconversion to IgG (9.7%), IgM (9.7%) and IgA (9.7%). Of the individuals 58.1% (18/31) showed synchronous seroconversion to S glycoprotein, RBD and N protein, whereas singular seroconversion to N protein or S glycoprotein was seen in 16.1% of individuals for each.
Longitudinal analysis across sequential samples highlighted the rapid decline in the IgM and IgA response to all three antigens after the peak OD between 20 and 30 d POS for IgM and IgA, respectively (Fig. 1b and Extended Data Fig. 1a), as might be expected after an acute viral infection14,22,23,24. For some individuals sampled at timepoints >60 d POS, the IgM and IgA responses were approaching baseline (Fig. 2b and Extended Data Fig. 1a). In contrast, the IgG OD (as measured at 1:50 dilution) remained high in most individuals, even up to 94 d POS (Fig. 1b and Extended Data Fig. 1a). However, differences were apparent when patients were stratified by disease severity (Fig. 2b) and when half-maximal binding (EC50) was measured (see Neutralizing antibody responses to SARS-CoV-2; Fig. 4b–d).
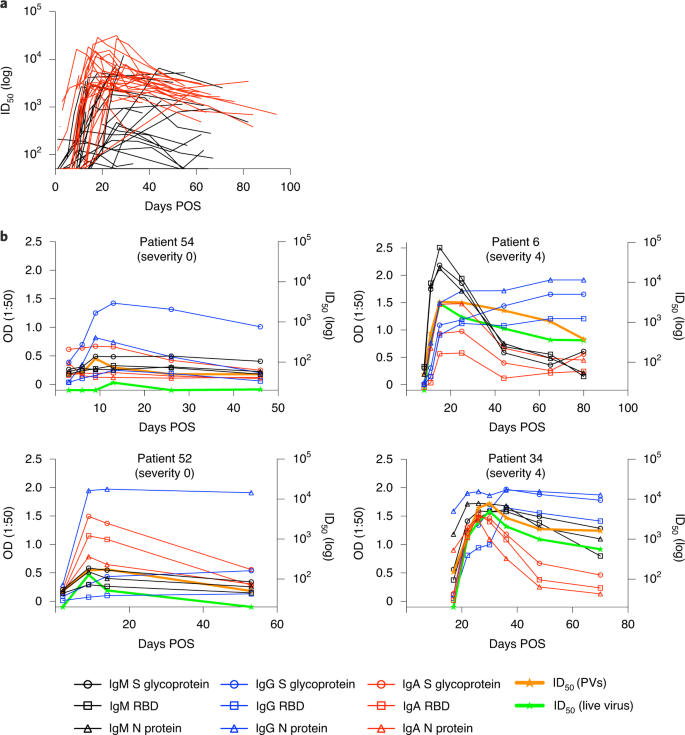
a, Neutralizing antibody ID50 changes related to days POS. ID50 was measured using HIV-1-based virus particles (PVs), pseudotyped with the S glycoprotein of SARS-CoV-2. Each line represents one individual (n = 65). Severities 0–3 are shown in black and severities 4/5 in red. Pre-COVID-19 healthy control samples did not show any neutralization at a 1:20 serum dilution (Supplementary Table 3). A subset of neutralization experiments (n = 25) was conducted twice, yielding similar results. ID50 values for the remaining samples were measured once. b, Example kinetics of antibody responses (IgM, IgA, IgG binding to S glycoprotein, RBD and N protein, and ID50 against PVs and wild-type virus) for four individuals during acute infection and the convalescent phase. Graphs show comparison between disease-rated severity 0 (left) and disease-rated severity 4 (right). The cut-off for the pseudovirus and wild-type virus neutralization assays are 1:50 and 1:20, respectively.
Neutralizing antibody responses to SARS-CoV-2
We next measured SARS-CoV-2 neutralization potency using a surrogate viral inhibition assay that uses human immunodeficiency virus type 1 (HIV-1)-based virus particles, pseudotyped with the S glycoprotein of SARS-CoV-2 (refs. 25,26) and a HeLa cell-line stably expressing the ACE-2 receptor. Increased neutralization potency was observed with increasing days POS (Fig. 2a), with each individual reaching a peak neutralization titre (range 98–32,000) on average 23.1 d POS (range 1–66 d) (Extended Data Fig. 1b). Two individuals (3.1%) did not develop a neutralizing antibody response (ID50 < 50) consistent with their lack of binding antibodies at the timepoints tested (<8 d POS). At peak neutralization, 7.7% had low (50–200), 10.8% medium (201–500), 18.5% high (501–2,000) and 60.0% potent (2,001+) neutralizing titres. For serum samples collected after 65 d POS, the percentage of donors with potent neutralizing antibodies (ID50 2,001+) had reduced to 16.7% (Supplementary Table 3). ID50 values correlated well with IgG-, IgM- and IgA-binding OD values to all three antigens, S glycoprotein, RBD and N protein (Extended Data Fig. 3a), and the best fit (r2) was observed between ID50 and the OD for S glycoprotein IgA and S glycoprotein IgM. The average time to detectable neutralization was 14.3 d POS (range 3–59 d). At earlier timepoints, some individuals displayed neutralizing activity before an IgG response to S glycoprotein and RBD was detectable by ELISA (Extended Data Fig. 2c). This highlights the capacity of S glycoprotein- and RBD-specific IgM and IgA to facilitate neutralization in acute infection in the absence of measurable IgG27.
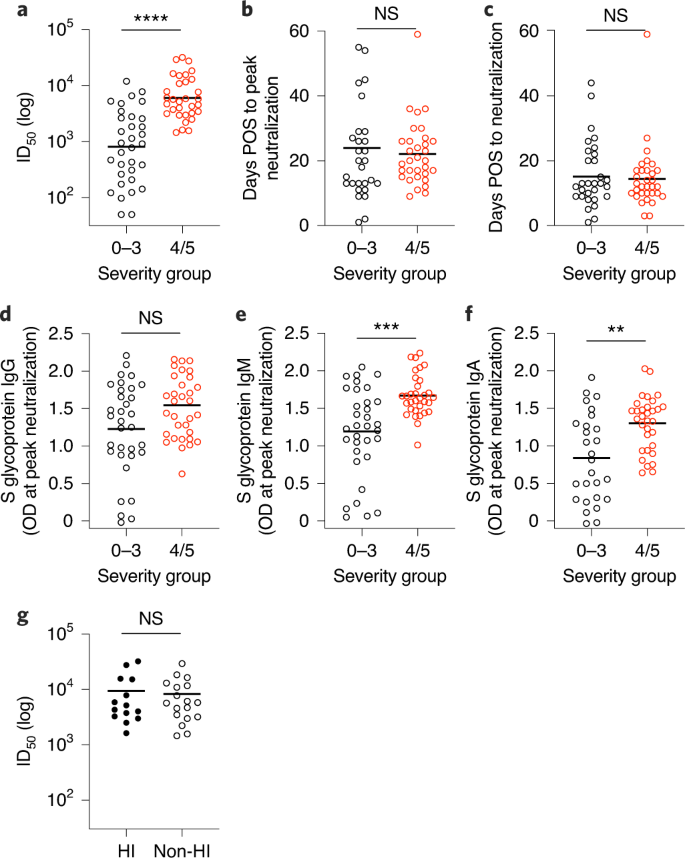
To determine how disease severity impacts neutralizing antibody titres, we compared the peak ID50 values between individuals with 0–3 and 4/5 disease severity (Fig. 3). Although the magnitude of the neutralizing antibody response at peak neutralization was significantly higher in the severity 4/5 group (Fig. 3a), the mean time taken to measure detectable neutralizing antibody titres (Fig. 3c) and the mean time to reach peak neutralization (Fig. 3b) did not differ between the two groups, suggesting that disease severity enhances the magnitude of the neutralizing antibody response but does not alter the kinetics. Comparison of the IgG, IgM and IgA OD values against S glycoprotein at peak neutralization showed significantly higher IgA and IgM ODs in the severity 4/5 group, but no significant difference was observed for IgG to S glycoprotein (Fig. 3d–f). This observation further highlights a potential role for IgA and IgM in neutralization27. Within the severity 4/5 group, a proportion of the patients was treated with immunomodulation for a persistent hyperinflammatory state characterized by fevers, markedly elevated C-reactive protein and ferritin, and multiorgan dysfunction. Despite an initial working hypothesis that antibody responses may differ as either a cause or a consequence of this phenotype, no difference in ID50 was observed between these individuals and the remainder of the severity 4/5 cases (Fig. 3g).
a–c, Comparison for individuals with disease severity 0–3 (n = 33 individuals) or 4/5 (n = 32 individuals) for peak ID50 of neutralization (a; P < 0.0001), the time POS to reach peak ID50 (b; P = 0.674) and the time POS to detect neutralizing activity (c; P = 0.9156). ID50 measured using HIV-1-based virus particles, pseudotyped with S glycoprotein of SARS-CoV-2. d–f, Comparison of OD values for individuals with 0–3 or 4/5 disease severity for IgG (d; P = 0.0635), IgM (e; P = 0.0003) and IgA (f; P = 0.0018) against S glycoprotein measured at peak ID50. g, Comparison of the peak ID50 value for individuals who were treated for hyperinflammation (HI; n = 14 individuals) or not treated (n = 18 individuals), and had 4/5 disease severity (P > 0.999). Statistical significance was measured using a Mann–Whitney two-sided test U-test. **P < 0.002, ***P < 0.0002, ****P < 0.0001. NS, not significant. The line represents the mean ID50 for each group.
Longevity of the neutralizing antibody response
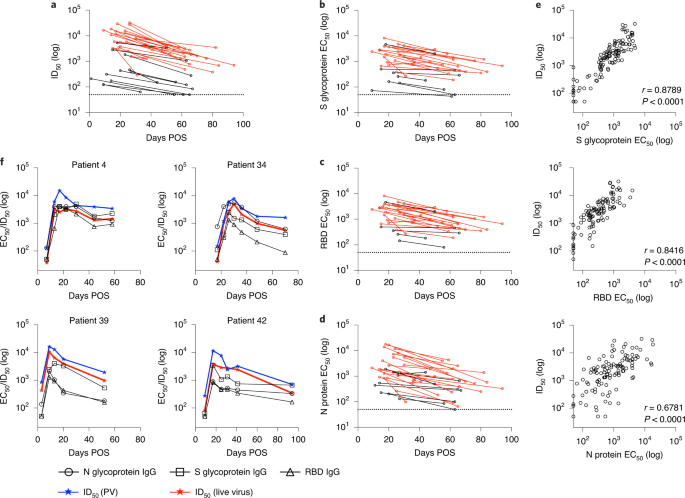
After the peak in neutralization, a waning in ID50 was detected in individuals sampled at >40 d POS. Comparison of the ID50 at peak neutralization and at the final timepoint collected showed a decrease in almost all cases (Fig. 4a). For some individuals with severity score 0, where peak neutralization was in the ID50 range 100–300, neutralization titres became undetectable (ID50 < 50) in the pseudotype neutralization assay at subsequent timepoints (Figs. 4a and 2b). For example, donors 52 and 54 both generated a low neutralizing antibody response (peak ID50 of 174 and 434, respectively) but no neutralization could be detected (at 1:50 dilution) against the pseudotyped virus 39 and 34 d after the peak ID50, respectively (Fig. 2b). To determine whether similar neutralization trends were observed with infectious wild-type virus, we selected a subset of individuals, representative of the range of neutralizing antibody responses observed using the pseudotyped virus, for further comparison. As shown by others, neutralization titres against authentic virus correlated very well (r2 = 0.9612, P < 0.0001) with those measured using the SARS-CoV-2 pseudovirus8,28,29 (Extended Data Fig. 3b) and the same trends in neutralization decline were observed for these selected donors (Figs. 2b and 4f). Neutralization of wild-type virus could also not be detected at 1:20 dilution in donors 52 and 54 at the final timepoints (Fig. 2b).
a, ID50 at peak neutralization (measured using HIV-1-based viral particles, pseudotyped with the S glycoprotein of SARS-CoV-2) is plotted with the donor-matched ID50 at the last timepoint from which serum was collected. Only individuals in whom the peak ID50 occurs before the last timepoint and the last timepoint is >30 d POS are included in this analysis (n = 35). The dotted line represents the cut-off for the pseudovirus neutralization assay. b–d, EC50 values for IgG binding to S glycoprotein (b), RBD (c) and N protein (d) were calculated at timepoints with peak ID50 and the final timepoint sera were collected. EC50 at peak neutralization is plotted with the donor-matched EC50 at the last timepoint sera were collected. Individuals with a disease severity 0–3 are shown in black and those with 4/5 in red. The dotted line represents the cut-off for EC50 measurement. The ELISAs to determine EC50 values were conducted once. e, Correlation of ID50 with IgG EC50 against S glycoprotein (r2 = 0.7924), RBD (r2 = 0.6781) and N protein (r2 = 0.4533) (Spearman’s correlation, r; a linear regression was used to calculate the goodness of fit, r2). f, Change in IgG EC50 measured against S glycoprotein, RBD and N protein, and ID50 using pseudovirus and wild-type virus over time for four example patients (all disease severity 4). The lowest dilution used for the pseudovirus and wild-type virus neutralization assays are 1:50 and 1:20, respectively.
To gain a more quantitative assessment of the longevity of the IgG-binding titres specific for S glycoprotein, RBD and N protein, EC50 values were measured by ELISA at peak neutralization and compared with the EC50 at the final timepoint collected. A stronger correlation was observed between ID50 and EC50 compared with the OD values (Fig. 4e and Extended Data Fig. 3). Similar to neutralization potency, a decrease in EC50 was observed within the follow-up period for S glycoprotein, RBD and N protein (Fig. 4b–d). For those whose neutralizing antibody titre decreased towards baseline, the EC50 for IgG to S glycoprotein and RBD also decreased in a similar manner. Finally, to determine whether the reduction in IgG titres might plateau, EC50 values were measured for all timepoints for four representative individuals, who had multiple samples collected in the convalescent phase (Fig. 4f). A steady decline in neutralization was accompanied by a decline in IgG EC50 to all antigens within the time window studied. Further assessment of antibody binding and neutralizing titres in samples collected >94 d POS will be essential to fully determine the longevity of the neutralizing antibody response.
Antibody responses in a HCW cohort
To gain further understanding of antibody responses in SARS-CoV-2 infection, we next analysed sequential serum samples from 31 seropositive (as determined by an IgG response to both N protein and S glycoprotein)6 HCWs from GSTFT. Antibody responses in these individuals are probably more akin to those from individuals who were never hospitalized. Sera were collected every 1–2 weeks from March 2020 to June 2020. Acute infection, as determined by rtqPCR, was not measured routinely, but symptoms relating to COVID-19 were recorded.
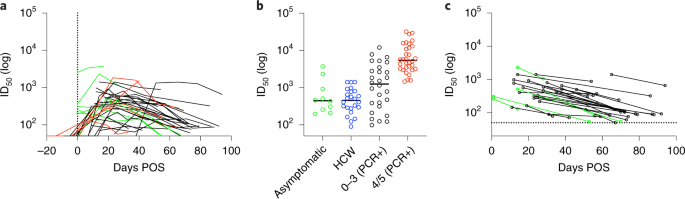
IgG and IgM binding to S glycoprotein, RBD and N protein by ELISA and pseudovirus neutralization titres were measured over time (Fig. 5a and Extended Data Fig. 4a). Similar to the patient cohort, ID50 values correlated with the OD values for IgG and IgM against S glycoprotein and RBD (Extended Data Fig. 4b). However, in contrast, the IgM and IgG responses to N protein in HCWs correlated poorly (r2 = 0.030 and 0.381, respectively) (Extended Data Fig. 4b). Comparison of the peak ID50 between asymptomatic individuals (6/31) and symptomatic HCWs (25/31) showed a very similar mean peak ID50. In contrast, both groups had lower mean peak ID50 values compared with hospitalized individuals in the 0–3 and 4/5 severity groups (Fig. 5b). Importantly, some asymptomatic individuals generated neutralization titres >1,000. Similar to the cohort with confirmed SARS-CoV-2 infection, a decline in ID50 was observed after peak neutralization. For many individuals with peak ID50 in the 100–500 range, neutralization was approaching baseline after 50 d POS (Fig. 5c). As the mean peak ID50 was lower in the HCW cohort, the decline in neutralizing antibody titres towards baseline was more frequent compared with the patient cohort.
a, ID50 values plotted against the time POS at which sera were collected. Each line represents one individual (n = 37). ID50 was measured using HIV-1-based viral particles pseudotyped with the S glycoprotein of SARS-CoV-2. Asymptomatic individuals are shown in green, symptomatic individuals in black and HCWs with PCR-confirmed SARS-CoV-2 infection in red (for comparison). The vertical dotted line represents day 0 POS. Of seropositive individuals, 80.6% (25/31) recorded COVID-19-compatible symptoms (including fever, cough and anosmia) since 1 February 2020, and 19.4% (6/31) reported none. A subset of neutralization experiments (n = 10) were conducted twice, yielding similar results. ID50 values for the remaining samples were measured once. b, Comparison of the peak ID50 between asymptomatic individuals (n = 10, includes 7 HCW and 3 hospital patients), HCWs (n = 24 symptomatic HCWs with no PCR test), and PCR+ individuals with severity either 0–3 (n = 28) or 4/5 (n = 32). The two PCR+ individuals, who were sampled at early timepoints (<8 d POS) and did not seroconvert, were not included in this analysis. c, ID50 at peak neutralization is plotted with the donor-matched ID50 at the last timepoint sera were collected. The dotted line represents the cut-off (1:50) for the SARS-CoV-2-pseudovirus neutralization assay. Asymptomatic donors are shown in green and symptomatic donors in black.
Discussion
The sequential serum samples collected up to 94 d POS allowed evaluation of the kinetics and longevity of the neutralizing antibody response in much greater detail than has hitherto been possible. As exemplified by Figs. 2b and 4f, the kinetics of the antibody response in SARS-CoV-2 infection is typical of an acute viral infection14,22,23,24. The peak in neutralizing antibodies arises due to a rapid production of short-lived plasmablasts30 that secrete high titres of antibodies; this is subsequently followed by a decline in virus-specific antibodies as these cells die. We observed a wide range of peak neutralizing antibody titres (98–32,000), similar to other cross-sectional cohorts21,4,31, and disease severity was associated with higher neutralizing antibody titres. It is not clear yet why neutralizing antibody responses correlate with disease severity32. A higher viral load may lead to more severe disease and generate a stronger antibody response through increased levels of viral antigen. Alternatively, antibodies could have a causative role in disease severity, although there is currently no evidence for antibody-dependent enhancement in COVID-1933.
Comparison of the peak ID50 for each individual, and ID50 at the final timepoint collected, showed a decline in neutralizing titres regardless of disease severity. The decline in neutralizing antibodies was mirrored in the reduction in IgG-binding titres (EC50) to S glycoprotein and RBD and also IgM and IgA binding to S glycoprotein and RBD (OD values) for the PCR+ cohort (Fig. 4b). For some individuals with peak ID50 in the 100–300 range, neutralizing titres were at, or below, the level of detection (ID50 < 50) after only ~50 d from the measured peak of neutralization (although IgG binding to N protein, S glycoprotein and RBD was still detected). This trend was also seen in the HCW cohort, and reveals that, in some individuals, SARS-CoV-2 infection generates only a transient neutralizing antibody response that rapidly wanes. For most individuals with peak ID50 titres >4,000, despite a decline in neutralizing antibody titres ranging from 2-fold to 23-fold over an 18- to 65-d period, neutralizing antibody titres remain in the 1,000–3,500 range at the final timepoint. Although the lowest serum dilution used in the pseudovirus neutralization assay is relatively high (1:50), donors who lacked detectable neutralization also showed no neutralizing activity against wild-type virus at 1:20.
The magnitude of the following decline in neutralizing antibody titres reported here is similar to that observed in several newly reported pre-prints12,13,20,31,34,35,36,37,38,39. In these studies, similar to our observations, those who generated a high neutralizing antibody titre still had high neutralizing antibody titres regardless of the initial decline12,13,31,35,39 and, for those with lower disease severity, two studies reported 7/34 (ref. 12) and 5/80 (ref. 31) individuals with a decline to undetectable neutralizing antibodies (ID50 < 20 or ID50 < 50, respectively) at the last timepoint studied. In contrast, several studies report a sustained antibody response in the first 3 months after SARS-CoV-2 infection, but these studies report changes in binding antibodies only34,37,38. Although binding titres to S glycoprotein and RBD correlate with neutralizing antibody titres, this difference in antibody measurement may account for the differences in kinetics described. Further follow-up in these longitudinal cohorts is required to determine whether the neutralizing antibody decline will continue on a downward trajectory or whether the neutralizing antibody titres will plateau to a steady state, facilitated through their production by long-lived plasma cells. Importantly, class-switched IgG memory B cells against S glycoprotein and RBD have been detected in blood of COVID-19 patients, showing that memory responses are generated during infection that have the potential to be activated to rapidly produce neutralizing antibodies on re-exposure to SARS-CoV-2 and prevent infection and/or disease8,29,40,41. Indeed, highly potent neutralizing monoclonal antibodies with protective capacity have been isolated from memory B cells of both symptomatic and asymptomatic individuals8,10,42.
The longevity of neutralizing antibody responses to other human coronaviruses has been previously studied14,15,16,17. In contrast to SARS-CoV-2 infection, SARS-CoV infection typically caused more severe disease and asymptomatic, low-severity cases were less common43. The neutralizing antibody response after SARS-CoV infection in a cohort of hospitalized patients peaked around day 30 (average titre 1:590)16 and a general waning of the binding IgG and neutralizing antibodies followed during the 3-year follow-up. Low neutralizing antibody titres of 1:10 were detected in 17/18 individuals after 540 d16. In a second study, low neutralizing antibody titres (mean 1:28) were still detected up to 36 months post-infection in 89% of individuals19. The lower neutralizing antibody responses in the 0–3 disease severity cases in our two cohorts may reflect more the immune response to endemic seasonal coronaviruses (that is, those associated with the common cold), which has also been reported to be more transient and where reinfections do occur2,18. For example, individuals experimentally infected with endemic α-coronavirus 229E, generated high antibody titres after 2 weeks, which rapidly declined in the following 11 weeks and, by 1 year, the mean antibody titres had reduced further14. Subsequent virus challenge led to reinfection (as determined by virus shedding), yet individuals showed no cold symptoms14.
The neutralizing antibody titre required for protection from reinfection and/or disease in humans is not yet understood. Neutralizing monoclonal antibodies isolated from SARS-CoV-2-infected individuals can protect from disease in animal challenge models in a dose-dependent manner, highlighting neutralizing antibodies as a correlate of protection9,10,11. SARS-CoV-2-infected rhesus macaques, which developed neutralizing antibody titres of ~100 (range 83–197), showed no clinical signs of illness when challenged 35 d after primary infection44. However, virus was still detected in nasal swabs, albeit 5 log lower than in primary infection, suggesting immunological control rather than sterilizing immunity. In contrast, a second study showed no detectable virus after re-challenge with neutralizing antibody titres in the 8–20 range45. Many current COVID-19 vaccine efforts focus on eliciting a robust neutralizing antibody response to provide protection from infection. Our observation that neutralizing antibody titres decline to low levels after low-severity disease suggests that vaccines should aim to elicit titres similar to those generated by severe disease and boosting may be required to maintain neutralizing antibody titres. The first results from phase I clinical trials showed peak median neutralizing antibody titres of 654 and 3,906 after two doses of a messenger RNA vaccine encoding S glycoprotein of SARS-CoV-2 (mRNA-1273) by Moderna46 and two doses of a recombinant nanoparticle S glycoprotein vaccine by Novavax47, respectively. Vaccine challenge studies in macaques can give some limited insights into neutralizing antibody titres required for protection from reinfection48,49,50,51. A DNA vaccine encoding S protein of SARS-CoV-2 generated neutralizing antibody titres between 100 and 200, which strongly correlated with a lowering of the viral load (up to 3 log)51. Further, vaccine mRNA-1273 generated geometric mean titres of 3,481 in macaques, which were shown to prevent viral replication in the upper and lower respiratory tracts52. The role that T cell responses generated through either infection53 or vaccination play in controlling disease cannot be discounted in these studies, and further definition of the correlates and longevity of vaccine-induced protection is needed. Taken together, despite the lower neutralizing antibody titres measured at the latest timepoints in some individuals, neutralizing antibody titres may still be sufficient to provide protection from COVID-19 for a period of time. However, follow-up studies involving sequential PCR testing and serological analysis in these individuals will be critical for understanding the ability of neutralizing antibodies to protect from reinfection in humans.
In summary, using sequential samples from SARS-CoV-2-infected individuals collected up to 94 d POS, we demonstrate a typical antibody response after an acute viral infection where a peak response was detected 3–4 weeks post-infection, which then wanes. For those who develop a low neutralizing antibody response (ID50 100–300), titres can return to baseline over a relatively short period, whereas those who develop a robust neutralizing antibody response maintain titres >1,000 despite the initial decline. Further studies using samples collected from these individuals at extended timepoints are required to determine the longevity of the neutralizing antibody response as well as the neutralizing antibody threshold for protection from reinfection and/or disease.
Methods
Ethics
Surplus serum from patient biochemistry samples taken as part of routine care was retrieved at the point of being discarded, aliquoted, stored and linked to a limited clinical dataset by the direct care team, before anonymization. Work was undertaken in accordance with the UK Policy Framework for Health and Social Care Research and approved by the Risk and Assurance Committee at GSTFT. Serum was collected from consenting HCWs with expedited approval from the GSTFT R&D office, occupational health department and medical director.
Patient and sample origin
Some 269 individual venous serum samples, collected at St Thomas’ Hospital, London from 59 patients diagnosed as SARS-CoV-2 positive via RT–PCR, were obtained for serological analysis. Samples ranged from 1 d to 94 d after onset of self-reported symptoms or, in asymptomatic cases, days after a positive PCR result. Patient information is given in Supplementary Table 1.
HCW cohort
Sequential serum samples were collected every 1–2 weeks from HCWs at GSTFT between 13 March and 10 June 2020. Seropositivity to SARS-CoV-2 was determined using sera collected in April and early May 2020, using ELISA. Individuals were considered seropositive if sera (diluted 1:50) gave an OD for IgG against both N protein and S glycoprotein that was fourfold above the negative control sera6. Self-reported COVID-19-related symptoms were recorded by participants and days POS in seropositive individuals was determined using this information. For asymptomatic, seropositive individuals, days POS was defined from the first timepoint at which SARS-CoV-2 antibodies were detected. Six participants had infection confirmed by PCR and were included with the PCR+ hospitalized patients in the initial analysis. An additional 31 HCWs were found to be seropositive and formed the HCW cohort.
COVID-19 severity classification
The score, ranging from 0 to 5, was devised to mitigate underestimating disease severity in patients not for escalation above level one (ward-based) care. Patients diagnosed with COVID-19 were classified as follows:
(0) Asymptomatic or no requirement for supplemental oxygen;
(1) Requirement for supplemental oxygen (fraction of inspired oxygen (FiO2) < 0.4) for at least 12 h;
(2) Requirement for supplemental oxygen (FiO2 ≥ 0.4) for at least 12 h;
(3) Requirement for non-invasive ventilation/continuous positive airway pressure or proning or supplemental oxygen (FiO2 > 0.6) for at least 12 h, and not a candidate for escalation above level one (ward-based) care;
(4) Requirement for intubation and mechanical ventilation or supplemental oxygen (FiO2 > 0.8) and peripheral oxygen saturations <90% (with no history of type 2 respiratory failure (T2RF)) or <85% (with known T2RF) for at least 12 h;
(5) Requirement for ECMO.
Cell lines
HEK293 freestyle cells were obtained from Thermo Fisher Scientific (catalogue no. R79007). HEK293T/17 cells were obtained from the American Type Culture Collection (ATCC CRL-11268). HeLa cells stably expressing ACE-2 (HeLa-ACE-2) were obtained from J. Voss (Scripps)10. Vero-E6 cells were obtained from W. Barclay (Imperial College London). All cell lines tested negative for Mycoplasma. No authentication was performed. No commonly misidentified cell lines were used.
Protein expression
The N protein of SARS-CoV-2 comprised residues 48–365 with an N-terminal, uncleavable, hexahistidine tag. N protein was expressed in Escherichia coli using autoinducing medium for 7 h at 37 °C, and purified using immobilized metal affinity chromatography, size exclusion and heparin chromatography. N protein was obtained from L. James and J. Luptak at LMB, Cambridge.
S glycoprotein consisted of the pre-fusion S ectodomain (residues 1–1138) with a GGGG substitution at the furin cleavage site (amino acids 682–685), proline substitutions at amino acid positions 986 and 987, and an N-terminal T4 trimerization domain followed by a Strep-tag II (ref. 8). The protein was expressed in HEK293F cells (Invitrogen). One litre of cells (density of 1.5 × 106 cells ml−1) were transfected with 325 µg DNA using PEI-Max (1 mg ml−1, Polysciences) at a 1:3 ratio and supernatant was harvested after 7 d. The protein was purified using StrepTactinXT Superflow high-capacity 50% suspension according to the manufacturer’s protocol by gravity flow (IBA Life Sciences). The plasmid was obtained from P. Brouwer, M. van Gils and R. Sanders at the University of Amsterdam.
The RBD protein (encoded by residues 319–541) has the natural N-terminal signal peptide of S glycoprotein fused at the start of the RBD sequence and is joined to a C-terminal hexahistidine tag. RBD was expressed in HEK293F cells (Invitrogen). Then, 500 ml of cells (density of 1.5 × 106 cells ml−1) was transfected with 1,000 µg DNA using PEI-Max (1 mg ml−1, Polysciences) at a 1:3 ratio. Supernatant was harvested after 7 d and protein purified using nickel–nitrilotriacetic acid agarose beads. The RBD plasmid was obtained from F. Krammer at Mount Sinai University1.
ELISA protocol
ELISAs were carried out as previously described6,30,54. All sera/plasma were heat inactivated at 56 °C for 30 min before use. High-binding ELISA plates (Corning, 3690) were coated with antigen (N protein, S glycoprotein or RBD) at 3 µg ml−1 (25 µl per well) in phosphate-buffered serum (PBS), either overnight at 4 °C or for 2 h at 37 °C. Wells were washed with PBS-T (PBS with 0.05% Tween-20) and then blocked with 100 µl of 5% milk in PBS-T for 1 h at room temperature. The wells were emptied and serum diluted at 1:50 in milk was added and incubated for 2 h at room temperature. Control reagents included CR3009 (2 µg ml−1), CR3022 (0.2 µg ml−1), negative control plasma (1:25 dilution), positive control plasma (1:50) and blank wells. Wells were washed with PBS-T. Secondary antibody was added and incubated for 1 h at room temperature. IgM was detected using goat-anti-human-IgM-HRP (horseradish peroxidase) (1:1,000) (Sigma, catalogue no. A6907), IgG was detected using goat-anti-human-Fc-AP (alkaline phosphatase) (1:1,000) (Jackson, catalogue no. 109-055-098) and IgA was detected goat-anti-human-IgA-HRP (1:1,000) (Sigma, catalogue no. A0295). Wells were washed with PBS-T and either AP substrate (Sigma) was added and read at 405 nm (AP) or one-step 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Thermo Fisher Scientific) was added and quenched with 0.5 M H2S04 before reading at 450 nm (HRP).
ELISA measurements were performed in duplicate and the mean of the two values was used.
EC50 values were measured using a titration of serum starting at 1:50 and a fivefold dilution series. EC50 values were calculated using GraphPad Prism. Measurements were carried out in duplicate.
SARS-CoV-2-pseudotyped virus preparation
Pseudotyped HIV virus incorporating the S glycoprotein of SARS-Cov-2 was produced in a 10-cm dish seeded the day before with 3.5 × 106 HEK293T/17 cells in 10 ml of complete Dulbecco’s modified Eagle’s medium (DMEM-C, 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin) containing 10% (v:v) FBS, 100 IU ml−1 of penicillin and 100 μg ml−1 of streptomycin. Cells were transfected using 35 μg of PEI-Max (1 mg ml−1, Polysciences) with: 1,500 ng of HIV-luciferase plasmid, 1,000 ng of HIV 8.91 gag/pol plasmid and 900 ng of the SARS-CoV-2 S glycoprotein plasmid25,26. The medium was changed 18 h post-transfection and the supernatant was harvested 48 h post-transfection. Pseudotype virus was filtered through a 0.45-μm filter and stored at −80 °C until required.
Viral entry inhibition assay with SARS-CoV-2-pseudotyped virus
Neutralization assays were conducted as previously described54. Serial dilutions of serum samples (heat inactivated at 56 °C for 30 min) were prepared with DMEM (10% FBS and 1% penicillin–streptomycin) and incubated with pseudotype virus for 1 h at 37 °C in 96-well plates. Next, HeLa cells stably expressing the ACE-2 receptor (provided by J. Voss, Scripps Research) were added (12,500 cells per 50 µl per well) and the plates were left for 72 h. The infection level was assessed in lysed cells with the Bright-Glo luciferase kit (Promega), using a Victor X3 Multilabel Reader (Perkin Elmer). Measurements were performed in duplicate and duplicates used to calculate the ID50.
Virus strain and propagation
Vero-E6 (Cercopithecus aethiops-derived epithelial kidney cells, provided by W. Barclay, Imperial College London) cells were grown in DMEM (Gibco) supplemented with GlutaMAX, 10% FBS and 20 µg ml−1 of gentamicin, and incubated at 37 °C with 5% CO2. SARS-CoV-2 Strain England 2 (England 02/2020/407073) was obtained from Public Health England. The virus was propagated by infecting 60–70% confluent Vero-E6 cells in T75 flasks, at a multiplicity of infection of 0.005 in 3 ml of DMEM supplemented with GlutaMAX and 10% FBS. Cells were incubated for 1 h at 37 °C before adding 15 ml of the same medium. Supernatant was harvested 72 h post-infection after visible cytopathic effect, and filtered through a 0.22-µm filter to eliminate debris, aliquoted and stored at −80 °C. The infectious virus titre was also determined by plaque assay in Vero-E6 cells.
Live virus neutralization assay
Vero-E6 cells were seeded at a concentration of 20,000 cells per 100 μl per well in 96-well plates and allowed to adhere overnight. Serial dilutions of serum samples (heat inactivated at 56 °C for 30 min) were prepared with DMEM (2% FBS and 1% PBS) and incubated with authentic SARS-CoV-2 for 1 h at 37 °C. The medium was removed from the pre-plated Vero-E6 cells and the serum–virus mixtures were added to the Vero-E6 cells and incubated at 37 °C for 24 h. This virus/serum mixture was aspirated and each well was fixed with 150 µl of 4% formalin at room temperature for 30 min and then topped up to 300 µl using PBS. The cells were washed once with PBS and permeabilized with 0.1% Triton-X in PBS at room temperature for 15 min. The cells were washed twice with PBS and blocked using 3% milk in PBS at room temperature for 15 min. The blocking solution was removed and an N protein-specific monoclonal antibody (murinized-CR3009) was added at 2 µg ml−1 (diluted using 1% milk in PBS) at room temperature for 45 min. The cells were washed twice with PBS and horse anti-mouse-IgG-conjugated to HRP was added (1:2,000 in 1% milk in PBS, Cell Signaling Technology, catalogue no. S7076) at room temperature for 45 min. The cells were washed twice with PBS, developed using TMB substrate for 30 min and quenched using 2M H2SO4 before reading at 450 nm.
Measurements were performed in duplicate and the duplicates used to calculate the ID50.
Statistical analysis
Analyses were performed using GraphPad Prism v.8.4.2.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The source data generated during the current study are available as supplementary information.
References
- 1.
Amanat, F. et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. https://doi.org/10.1038/s41591-020-0913-5(2020).
- 2.
Gorse, G. J., Donovan, M. M. & Patel, G. B. Antibodies to coronaviruses are higher in older compared with younger adults and binding antibodies are more sensitive than neutralizing antibodies in identifying coronavirus-associated illnesses. J. Med. Virol. 92, 512–517 (2020).
- 3.
Long, Q. X. et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 26, 845–848 (2020).
- 4.
Luchsinger, L. L. et al. Serological assays estimate highly variable SARS-CoV-2 neutralizing antibody activity in recovered COVID19 patients. J. Clin. Microbiol. https://doi.org/10.1128/JCM.02005-20 (2020).
- 5.
Okba, N. M. A. et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 26, 1478–1488 (2020).
- 6.
Pickering, S. et al. Comparative assessment of multiple COVID-19 serological technologies supports continued evaluation of point-of-care lateral flow assays in hospital and community healthcare settings. PLoS Pathog. 16, e1008817 (2020).
- 7.
Prevost, J. et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 Spike. Cell Rep. Med. 1, 100126 (2020).
- 8.
Brouwer, P. J. M. et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science https://doi.org/10.1126/science.abc5902 (2020).
- 9.
Cao, Y. et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell https://doi.org/10.1016/j.cell.2020.05.025 (2020).
- 10.
Rogers, T. F. et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science https://doi.org/10.1126/science.abc7520 (2020).
- 11.
Shi, R. et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature https://doi.org/10.1038/s41586-020-2381-y (2020).
- 12.
Crawford, K. H. D. et al. Dynamics of neutralizing antibody titers in the months after SARS-CoV-2 infection. J. Infect. Dis. https://doi.org/10.1093/infdis/jiaa618 (2020).
- 13.
Long, Q. X. et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 26, 1200–1204 (2020).
- 14.
Callow, K. A., Parry, H. F., Sergeant, M. & Tyrrell, D. A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 105, 435–446 (1990).
- 15.
Kellam, P. & Barclay, W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J. Gen. Virol. https://doi.org/10.1099/jgv.0.001439 (2020).
- 16.
Mo, H. et al. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology 11, 49–53 (2006).
- 17.
Moore, J. P. & Klasse, P. J. SARS-CoV-2 vaccines: ‘Warp Speed’ needs mind melds not warped minds. J. Virol. https://doi.org/10.1128/JVI.01083-20 (2020).
- 18.
Edridge, A. W. D. et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. https://doi.org/10.1038/s41591-020-1083-1 (2020).
- 19.
Cao, W. C., Liu, W., Zhang, P. H., Zhang, F. & Richardus, J. H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 357, 1162–1163 (2007).
- 20.
Lee, W. T. et al. Neutralizing antibody responses in COVID-19 convalescent sera. Preprint at medRxiv https://doi.org/10.1101/2020.07.10.20150557 (2020).
- 21.
Wu, F. et al. Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in Shanghai, China. JAMA Intern. Med. https://doi.org/10.1001/jamainternmed.2020.4616 (2020).
- 22.
Wang, M. et al. Antibody dynamics of 2009 influenza A (H1N1) virus in infected patients and vaccinated people in China. PLoS ONE 6, e16809 (2011).
- 23.
Amanna, I. J., Carlson, N. E. & Slifka, M. K. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 357, 1903–1915 (2007).
- 24.
Choe, P. G. et al. MERS-CoV antibody responses 1 year after symptom onset, South Korea, 2015. Emerg. Infect. Dis. 23, 1079–1084 (2017).
- 25.
Grehan, K., Ferrara, F. & Temperton, N. An optimised method for the production of MERS-CoV spike expressing viral pseudotypes. MethodsX 2, 379–384 (2015).
- 26.
Thompson, C. et al. Neutralising antibodies to SARS coronavirus 2 in Scottish blood donors—a pilot study of the value of serology to determine population exposure. Preprint at medRxiv https://doi.org/10.1101/2020.04.13.20060467 (2020).
- 27.
Sterlin, D. et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Preprint at medRxiv https://doi.org/10.1101/2020.06.10.20126532 (2020).
- 28.
Schmidt, F. et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 217, https://doi.org/10.1084/jem.20201181 (2020).
- 29.
Ju, B. et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature https://doi.org/10.1038/s41586-020-2380-z (2020).
- 30.
Laing, A. G. et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. https://doi.org/10.1038/s41591-020-1038-6 (2020).
- 31.
Muecksch, F. et al. Longitudinal analysis of clinical serology assay performance and neutralising antibody levels in COVID19 convalescents. Preprint at medRxiv https://doi.org/10.1101/2020.08.05.20169128 (2020).
- 32.
Lee, N. et al. Anti-SARS-CoV IgG response in relation to disease severity of severe acute respiratory syndrome. J. Clin. Virol. 35, 179–184 (2006).
- 33.
Iwasaki, A. & Yang, Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 20, 339–341 (2020).
- 34.
Wajnberg, A. et al. SARS-CoV-2 infection induces robust, neutralizing antibody responses that are stable for at least three months. Preprint at medRxiv https://doi.org/10.1101/2020.07.14.20151126 (2020).
- 35.
Iyer, A. S. et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 5, eabe0367 (2020).
- 36.
Beaudoin-Bussières, G. et al. Decline of humoral responses against SARS-CoV-2 Spike in convalescent individuals. mBio 11, e02590–20 (2020).
- 37.
Isho, B. et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 5, eabe5511 (2020).
- 38.
Wu, J. et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Preprint at medRxiv https://doi.org/10.1101/2020.07.21.20159178 (2020).
- 39.
Wang, K. et al. Longitudinal dynamics of the neutralizing antibody response to SARS-CoV-2 infection. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa1143 (2020).
- 40.
Seydoux, E. et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity https://doi.org/10.1016/j.immuni.2020.06.001 (2020).
- 41.
Rodda, L. B. et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Preprint at medRxiv https://doi.org/10.1101/2020.08.11.20171843 (2020).
- 42.
Robbiani, D. F. et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature https://doi.org/10.1038/s41586-020-2456-9 (2020).
- 43.
Petersen, eK. M. et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. 20, e238–e244 (2020).
- 44.
Chandrashekar, A. et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science https://doi.org/10.1126/science.abc4776 (2020).
- 45.
Deng, W. et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science https://doi.org/10.1126/science.abc5343 (2020).
- 46.
Jackson, L. A. et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N. Engl. J. Med. https://doi.org/10.1056/NEJMoa2022483 (2020).
- 47.
Keech, C. et al. Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. https://doi.org/10.1056/NEJMoa2026920 (2020).
- 48.
Gao, Q. et al. Rapid development of an inactivated vaccine candidate for SARS-CoV-2. Science https://doi.org/10.1126/science.abc1932 (2020).
- 49.
Smith, T. R. F. et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 11, 2601 (2020).
- 50.
van Doremalen, N. et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature https://doi.org/10.1038/s41586-020-2608-y (2020).
- 51.
Yu, J. et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science https://doi.org/10.1126/science.abc6284 (2020).
- 52.
Corbett, K. S. et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. https://doi.org/10.1056/NEJMoa2024671 (2020).
- 53.
Sekine, T. et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell Press 183, 158–168 (2020).
- 54.
Carter, M. J. et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat. Med. https://doi.org/10.1038/s41591-020-1054-6 (2020).
Acknowledgements
King’s Together Rapid COVID-19 Call awards were received by M.H.M., K.J.D., S.J.D.N. and R.M.N. MRC Discovery award MC/PC/15068 was received by S.J.D.N., K.J.D. and M.H.M. J.S., S.P. and J.M.J.-G. received support from the Huo Family Foundation (M.H.M., S.J.D.N,. K.J.D., R.M.N., J.D.E. and M.S.-H.). A.W.S. and C.G. were supported by the MRC-KCL Doctoral Training Partnership in Biomedical Sciences (MR/N013700/1). G.B. and J.M.J.-G. were supported by the Wellcome Trust (grant no. 106223/Z/14/Z to M.H.M.). S.A. was supported by an MRC-KCL Doctoral Training Partnership in Biomedical Sciences industrial Collaborative Award in Science & Engineering (iCASE) in partnership with Orchard Therapeutics (MR/R015643/1). N.K. was supported by the Medical Research Council (MR/S023747/1 to M.H.M.). M.S.H. is supported by the National Institute for Health Research Clinician Scientist Award (CS-2016-16-011). The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research or the Department of Health and Social Care. S.P., H.D.W. and S.J.D.N. were supported by a Wellcome Trust Senior Fellowship (WT098049AIA). Fondation Dormeur, Vaduz provided funding for equipment (to K.J.D.). Development of SARS-CoV-2 reagents (RBD expression plasmid) was partially supported by the NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS; contract no. HHSN272201400008C). We thank F. Krammer for provision of the RBD expression plasmid, P. Brouwer, M. van Gils and R. Sanders (University of Amsterdam) for the S glycoprotein construct; L. James, J. Luptak and L. Kiss (LMB) for the provision of purified N protein; and J. Voss and D. Huang for providing the HeLa-ACE-2 cells. We thank L. Mccoy (UCL) for the provision of the murine CR3009 monoclonal antibody. We thank W. James (University of Oxford) for advice on microneutralization assays. We thank all patients and staff at St Thomas’ Hospital who participated in this study. We thank the COVID-19 core research team members, including O. Tijani, K. Brooks, M. Flanagan, R. Kaye, R. Williams, C. Blanco-Gil, H. Kerslake, A. Walters, R. Dakari, J. Squires, A. Stanton, S. Tripoli, A. Amon, I. Chow, O. Okolo and N. Lumlertgul.
Author information
Affiliations
Contributions
K.J.D., B.M., S.J.D.N., M.H.M. and J.D.E. designed the study. J.S., C.G., S.A., K.J.A.S., O.H., A.O’B., N.K., I.H. and K.J.D. performed ELISAs. J.S., C.G. and S.P. performed neutralization assays. H.W., C.K., R.M.N., M.J.L. and J.M.J.-G. prepared pseudovirus or wild-type virus. J.S., C.G., B.M., S.A., K.J.D., M.H.M. and S.J.D.N. analysed and interpreted the data. S.P., R.P.G., G.B., H.D.W. and A.W.S. curated hospital serum samples. B.M., K.B., A.P., M.K.I.T., L.O.C., G.O’H., E.M., S.D., G.N. and R.B. assisted in collection of samples from hospitalized patients. B.M., L.S., K.B., A.M., A.G., L.M., B.S., J.H., A.I.-B., G.A., A.P. and R.B. assisted in collection of samples from HCWs. G.O’H., E.M., S.D. and G.N. assisted in project administration. N.T. provided new reagents. K.J.D., M.H.M., S.J.D.N., B.M., J.D.E., J.S., C.G., S.P., R.P.G. and M.S.H. drafted the manuscript or substantially revised it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cross-sectional SARS-CoV-2 Ab responses.
a, IgM, IgA and IgG OD values against S glycoprotein, RBD and N protein are binned based on days post-onset of symptoms (POS) for 302 separate serum samples taken sequentially from 65 individuals. The data is grouped based on disease severity, 0–3 (black) and 4/5 (red). The mean OD value is shown for each time window for both the 0–3 (black line) and 4/5 (red line) severity groups. OD = optical density. b, ID50 measured using the SARS-CoV-2 pseudotype neutralization assay are binned based on days post-onset of symptoms (POS). The data is grouped based on disease severity, 0–3 (black) and 4/5 (red). The mean ID50 values are shown for each time window for both the 0–3 (black line) and 4/5 (red line) severity groups. The ELISA assays were conducted once. A subset of neutralization experiments (n=25) were conducted twice yielding similar results.
Extended Data Fig. 2 Antibody isotype and specificity at the timepoint for which an individual became seropositive.
a, A cumulative frequency analysis describing the seroconversion in the hospitalized cohort. Graph shows the percentage of individuals in the cohort that become IgM, IgA or IgG seropositive against S glycoprotein, RBD and N protein each day POS. b, Patterns of seroconversion based on antigen and antibody isotype for a subset of donors from whom sera was collected over sequential early time points (<14 days POS, n=31). For individuals with at least 2 sera collected <14 days POS, the first antigen(s) and antibody isotype(s) that gave an OD that was 4-fold above background is reported. c) Pseudovirus neutralization and ELISA binding at early time points POS for a subset of individuals (n=15) where there were measurable neutralization titres but IgG binding to S was low (OD <0.2) and IgM and/or IgA to S is higher (OD >0.4). Each bar shows data for one patient only.
Extended Data Fig. 3 Correlations between OD values and ID50.
a, Correlation of ID50 measured against SARS-CoV-2 pseudovirus with IgG (blue), IgM (black), IgA (red) OD values (at 1:50) against S glycoprotein, RBD and N protein (Spearman correlation, r). b, Correlation of ID50 measured using wild-type virus and ID50 measured using pseudovirus neutralization assays (Spearman correlation, r). A linear regression was used to calculate the goodness of fit (r2). The dotted lines represent the lowest serum dilution used in each assay. The lowest dilution used for the pseudovirus and wild-type virus neutralization assays are 1:50 and 1:20, respectively.
Extended Data Fig. 4 Ab responses in a healthcare worker cohort.
a, Ab responses (IgG and IgM to S glycoprotein, RBD and N protein) over time. Each line represents one individual. Asymptomatic individuals shown in green, symptomatic individuals shown in black and PCR+ HCW shown in red (for comparison). The dotted line represents day 0 POSs. b, Correlation of ID50 with IgG and IgM OD values against S glycoprotein and N protein (Spearman correlation, r). A linear regression was used to calculate the goodness of fit (r2). The ELISA assays were conducted once. A subset of neutralization experiments were conducted twice yielding similar results.
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Supplementary Data
OD, ID50 and EC50 values for each serum sample used in the study.
Cite this article
Seow, J., Graham, C., Merrick, B. et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol (2020). https://doi.org/10.1038/s41564-020-00813-8
Received
Accepted
Published
No hay comentarios:
Publicar un comentario